Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
Physical Address
304 North Cardinal St.
Dorchester Center, MA 02124
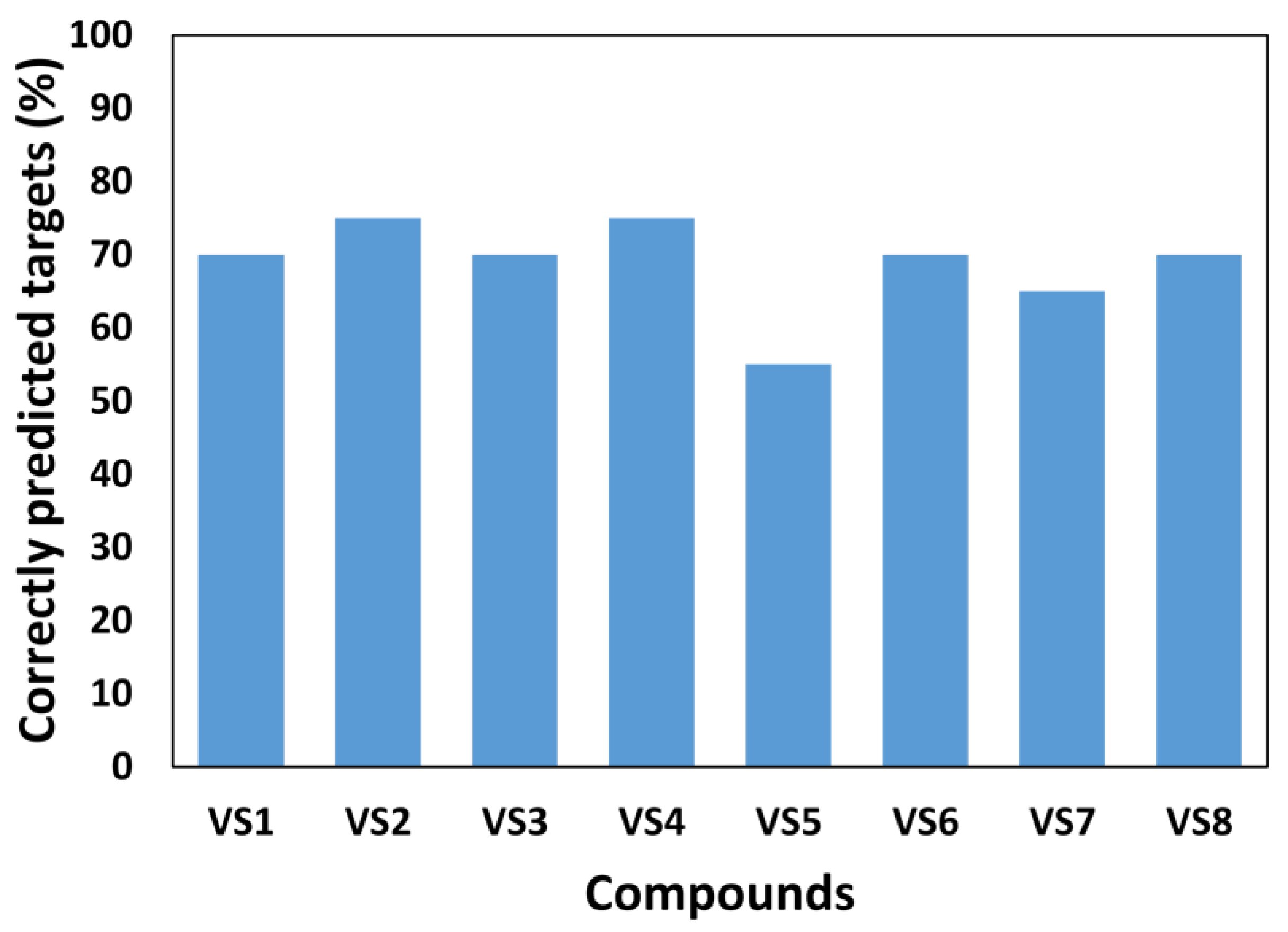

Protein kinase research presents an opportunity to explore molecular targets in the body to treat diseases like cancer and autoimmune disorders. These enzymes have the potential to bind to cellular sites and inhibit dysfunctional behavior, such as the overproduction of cancerous cells and tumor formation.
With the vast combination of possible kinase and cell structures, scientists are turning to Artificial Intelligence (AI) in order to predict and create a model of which pairing could have a therapeutic effect.
A team of researchers have developed KinasePred, a computational tool for small-molecule kinase target prediction. They published the details of their project in the International Journal of Molecular Sciences.
The authors include researchers collaborating with the Sbarro Health Research Organization (SHRO), under the direction of Antonio Giordano, M.D., Ph.D., Professor at Temple University, with scientists from the University of Pisa, and other research organizations in Italy. This AI-based workflow is able to predict kinase activity, to gain insights into molecular target interactions and identify combinations with the potential to treat cancer.
The study, led by Dr. Miriana de Stefano, Department of Pharmacy, University of Pisa, presents an advanced computational tool designed to strengthen the prediction of kinase interactions with small molecules.
KinasePred is an example of a data-dependent computational tool developed to solve a particular problem—the selection of kinase inhibitors. This is achieved by applying a predictive model which uses the molecular basis of binding and selectivity of kinases.
KinasePred uses Machine Learning (ML) and AI to make accurate predictions as well as explain the molecular characteristics that facilitate the interactions. The researchers hope the tool will lead to more accurate predictions using new representations of the molecules and different machine learning methods, and provide a more comprehensive knowledge of kinase interactions.
These advancements are critical to identifying and minimizing off-target effects, ultimately enhancing the safety and selectivity of therapeutic agents.
More information:
Miriana Di Stefano et al, KinasePred: A Computational Tool for Small-Molecule Kinase Target Prediction, International Journal of Molecular Sciences (2025). DOI: 10.3390/ijms26052157
Provided by
Sbarro Health Research Organization (SHRO)
Citation:
Computational model enhances protein kinase target prediction for therapies (2025, March 11)
retrieved 12 March 2025
from https://phys.org/news/2025-03-protein-kinase-therapies.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.